This article shows the ADE Discovery Browser can contribute to the visibility of adverse drug events (ADEs) – and thereby drug safety – by an end-to-end qualitative text mining of ADEs associated with a medication like the COVID-19 vaccines.
The outbreak of a global pandemic revealed the need for quick but also safe development of COVID-19 vaccines. Next to the practical complexity of developing and manufacturing such vaccines, the amount of research accompanying the drug development constituted a challenge by itself. Biomedical literature hosts research findings and case reports of crucial and potential hidden information such as adverse drug events (ADEs) associated with a certain medication consumption. Recently developed COVID-19 vaccines were observed to cause mild ADEs such as fever, redness at the injection site, headache, diarrhoea, muscle pain and also several severe ADEs such as anaphylaxis allergic reactions, thrombocytopenia, bell’s palsy etc. in certain groups of people or individuals. Since the COVID-19 vaccines are being produced and administered, the amount of available literature related to ADEs associated with COVID-19 vaccines have been increasing just like the amount of available biomedical literature in any other topic.
In order to contribute to the visibility of ADEs and thereby drug safety, our goal was to perform end-to-end qualitative text mining of ADEs associated with a medication from the life sciences literature database, PubMed, and to make it possible to interactively discover the ADEs, DRUGs and their causal and semantic relationships. The eventual goal of the ADE Discovery Browser is to provide a system to pharmacovigilance experts where they can easily make use of and evaluate the retrieved information about ADEs from the scientific literature. To achieve that, we performed 2 tasks: named entity recognition (NER) and relation extraction (RE). For each task, we fine-tuned transformers based pre-trained language models (PLM): a generic domain model DistilBERT and an in-domain model PubMedBERT.
What is ADE?
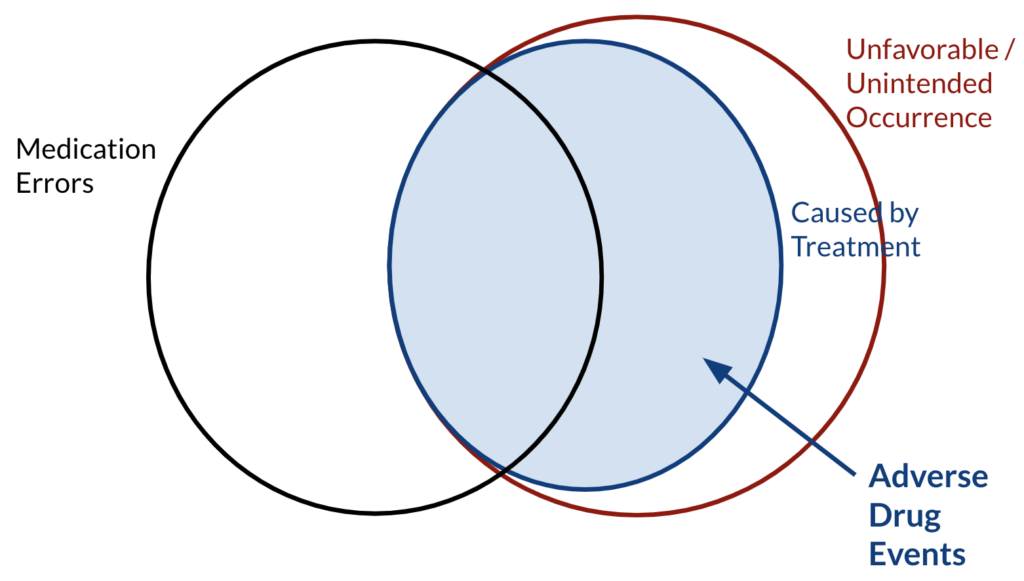
First, let’s leave out the word “drug“ and focus on “adverse events“ only. Adverse events are any unwanted and unfavourable occurrence that is temporarily associated with the use of a medical product. In this sense, “occurrence“ can be a sign or a symptom, “medical product“ can be a drug, medical device or a vaccination and “associated with“ does not necessarily imply causation. There exists a subset in this set of undesired occurrences which are caused by treatment, and some of these undesired occurrences that are caused by treatment happen due to medication errors. Medication errors occur either during prescribing such as failure to adjust with different drugs, or administration, such as overdosing or having allergic reactions etc..

![Figure 2: ADRs in a Venn Diagram [1] ADRs in a Venn Diagram](https://www.inovex.de/wp-content/uploads/adr-1024x585.png)
Natural Language Processing for ADE Extraction
In the biomedical domain, there are several text based resources like electronic health records, biomedical literature, medical blog websites, patient surveys, laboratory reports etc.. Having such data resources more and more electronically available, brings the idea if natural language processing (NLP) techniques can be helpful to extract ADEs automatically from such text-based resources because they potentially include hidden ADE cases, and the more ADEs known, the more they can be prevented. In order to perform such information retrieval,
- automatic recognition of DRUG and ADE named entities in a given text
- automatic extraction of DRUG-ADE relations between the two target entities
needs to be handled using NLP techniques.
Let’s say the following sentence is from one of the text-based resources. The first step is to identify the ADE and DRUG mention spans in the sentence in order to relate the observation of an ADE to a drug as in the following example.

Once we have all the ADE and DRUG entity spans identified, we have a set of ADEs and a set of DRUGs and we do not know yet which of those are actually in a causal relationship with which of those. Thereafter, the second step is to identify the semantic relationships between the entities in order to assign a causal relationship. Eventually, amoxicillin is in a causal relation with anaphylactic reaction, pruritis and facial erythema edema, and bactrim is in a causal relation with pruritis, facial erythema edema and anaphylactic exposure.
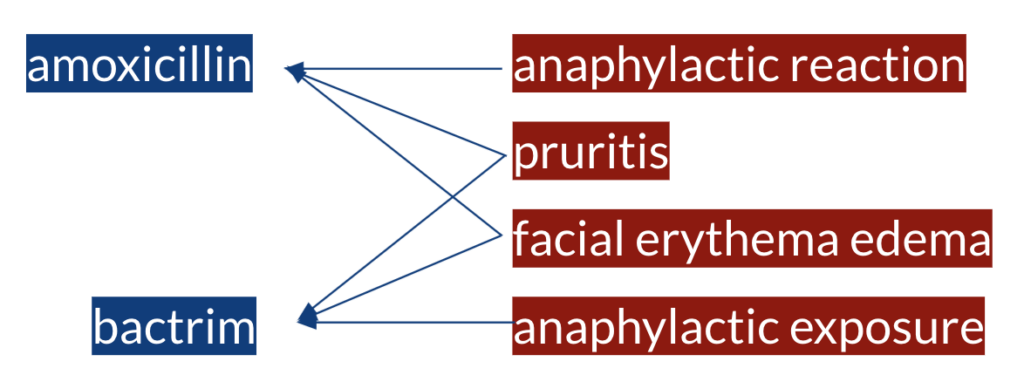
As can be seen from the example above, some of the challenges of the task are to have multiple entities of same or different types in a sentence, entities with multiple words and that not all the entity pairs mentioned in a sentence are in a relation. These challenges can be approached using NLP techniques and following the tasks such as named entity recognition and relation extraction.
Our Approach, Data and Results
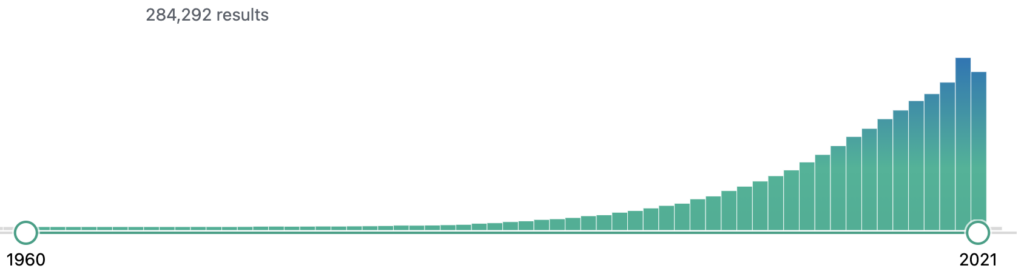
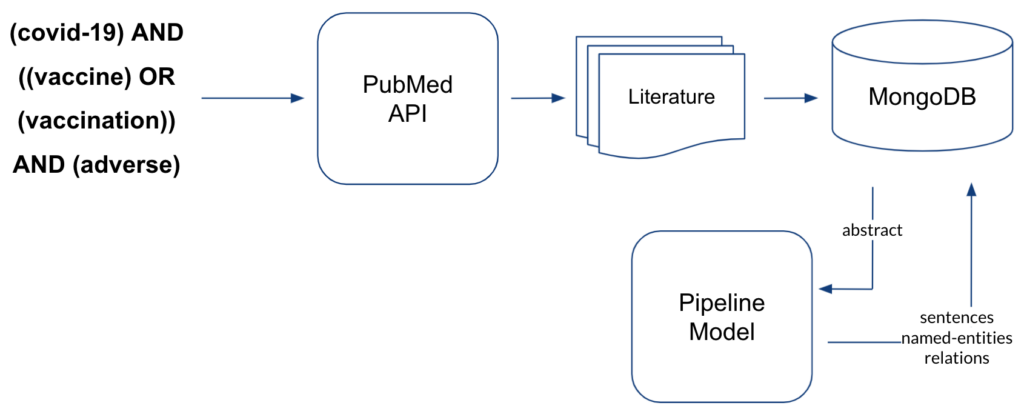
Our approach was to develop an end-to-end solution for interactive discovery of ADEs from the scientific literature in order to provide pharmacovigilance experts or researchers a system with easy information gathering of ADEs from the published research papers. PubMed is a database of around 30M life sciences literature. The amount of available biomedical literature in PubMed is increasing exponentially year by year, as shown in Figure 6, which makes it impossible for any researcher to stay up-to-date by reading and getting insights from the literature. Moreover, the literature potentially hosts information about common and rare ADEs that are observed and published as research findings or case reports of adverse events associated with a certain drug consumption.
Such common and rare ADEs can be retrieved from the literature by applying the below mentioned information retrieval techniques and our approach is illustrated in Figure 7.
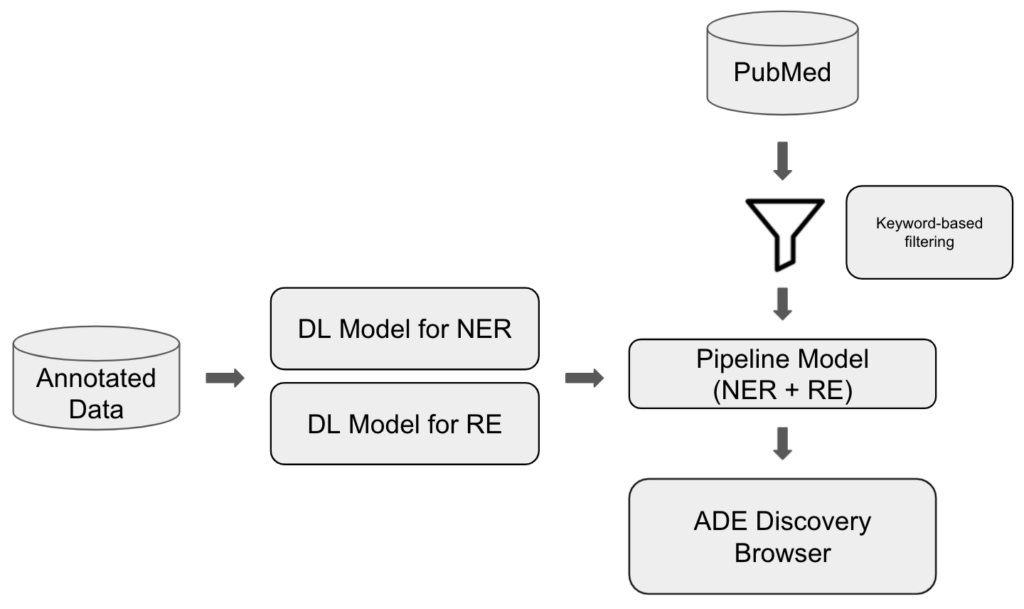
We trained two deep learning (DL) models making use of transformers-based pre-trained language models from the Hugging Face library: one performing the named entity recognition (NER) of DRUGs and ADEs and one performing the relation extraction (RE) whether a DRUG-ADE pair is related to each other in a sentence or not. We fine-tuned the models using the annotated data, and then pipelined them in a way that the output of NER can be fed into RE as input so that we can process sequential evaluation. Then, we have a set of literature filtered and downloaded from PubMed and we apply this literature to the pipeline model and visualise the retrieved information in the ADE Discovery Browser. We worked with 2 annotated datasets for fine-tuning:
- 2018 n2c2 ADE and Medication Extraction Challenge dataset [5]
- ADE corpus [6]
To the best of our knowledge, the n2c2 dataset is one of the latest available annotated datasets which includes 1038 sentences with at least one ADE entity annotation out of 505 electronic health records. The ADE corpus was the first published labeled data for this task in 2012 by filtering 3000 case reports from PubMed and annotating the relevant sentences which includes 4272 sentences with at least one ADE annotation. The language domain of the ADE corpus is more in the medical context than the n2c2 dataset. The data resource being the biomedical literature instead of spontaneously written clinical records, ADE corpus has more specific domain words and includes more thought language. The big difference between the n2c2 dataset and the ADE corpus was that the n2c2 dataset also involves generic terms annotated as named entity. Such generic terms can be simply mentions of
painkiller, antibiotics
for drugs instead of specifically mentioning which kind of painkiller or antibiotics it was, or mentions of
overdose, drug reaction, allergic reaction
instead of specifying which kind of adverse event was observed. The problem is that the named entity is expected to be a more specific definition of DRUG or ADE. The n2c2 dataset also has an inconsistency problem. An observed sample was different ways of annotating rash observation as an ADE:
rash, developed a drug rash, developed papular rash over posterior thighs and perirectal area, red rash on arms and face, severe rash
ADE Corpus was able to successfully differentiate such named entities and includes more specific words for DRUG or ADE mentions. Therefore, we used ADE Corpus to evaluate the quality of the n2c2 dataset to introduce better quality labeled data and to introduce more data samples. For those reasons, we used the ADE Corpus together with the n2c2 dataset for advancing the named entity recognition while using only the n2c2 dataset for both named-entity recognition and relation extraction.
Named-Entity Recognition
Biomedical NER has been quite important in identifying specific biomedical entity types from unstructured text, and acts as a basis for many biomedical text analysis tasks. We approached NER as a sequential token classification problem and performed Beginning-Inside-Outside (BIO) tagging of the annotated corpora in order to fine-tune the PLMs. By using PLMs and the ADE corpus, we incorporated outside knowledge and a good language representation to the n2c2 dataset. The biggest motivation for using PLMs was to make use of state-of-the-art contextual representations and observe how much they can help the sequential token classification task for ADE extraction.
We performed entity level evaluation and considered F1 score as the main comparison metric. We observed the best F1 score for ADE entity span recognition, fine-tuning the PubMedBERT model and using the combined datasets. This setting indeed performed better than a previously published machine learning model using CRF and is comparable to a deep learning model of Bi-LSTM and CRF layers [7]. Therefore, we used the fine-tuned PubMedBERT model using the combined datasets for the pipeline processing later on.
Relation Extraction
Biomedical RE also has been an attention catching research topic in identifying semantic relations from biomedical text documents. RE relies on the two extracted entities and the sentence those entities were used. We approached RE as a sentence classification problem and performed entity masking of the target entities in the input sentence when fine-tuning the PLMs.
For entity masking, we replaced the actual tokens of the target entities with special tokens such as [ADE] and [DRUG] [8]. This prevents the model from learning the specific lexical items, and is useful for us because we want to keep discovering new relations depending on the context of the sentence and not on the tokens that make up the entity.
Usage of contextual representation is useful for RE tasks, because the identification of such relations heavily relies on the context the entities were used in. We can easily observe in the datasets that DRUG-ADE semantic relations in the sentence exist with phrases like:
occur .. after, induced, associated with, developed, suffered from, caused in, concluded to etc.
On the other side, we did not consider extracting relations across sentences because we decided the input sequence to be a single sentence at a time. Therefore, our model was only able to classify relations within the same sentence, and not across sentences.
We fine-tuned two different PLMs for RE, i.e. DistilBERT and PubMedBERT. While fine-tuning the DistilBERT model, we obtained comparable results for DRUG-ADE relation classification to a previously published classical model based on Support Vector Machines (SVM) [7]. However, fine-tuning the PubMedBERT model for sentence classification was not successful. Therefore, we used the fine-tuned DistilBERT model for the pipeline processing later on.
Information Retrieval
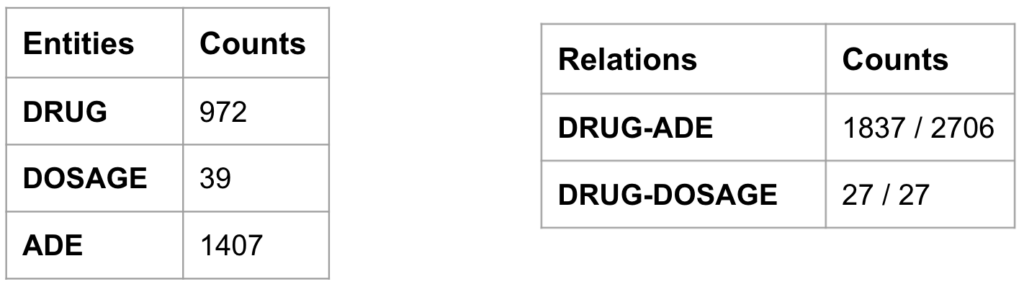
Once we had the NER and RE models performing just as good as the best team’s models and the pipeline processing in place, we could perform the information retrieval on biomedical literature to uncover the hidden and rare ADEs associated with COVID-19 vaccines as illustrated in Figure 8. With the search query mentioned in the below figure, we downloaded 1321 abstracts from PubMed which potentially talk about the ADEs after receiving COVID-19 vaccines. After processing the abstracts through the pipeline model, we stored the sentences, extracted named entities and the classified relations in the MongoDB. From these abstracts, we retrieved 972 DRUG entity mentions, 1407 ADE entity mentions, and 1837 sentences with DRUG-ADE relations which contain both target entities as summarised in Table 1.
Application Development
Finally, we implemented an application with user interaction in two different scenarios for the interactive discovery of ADEs:
- a user input paragraph can be processed through the pipeline and the user can visualise the entities highlighted in the paragraph together with a table of the relations,
- a user can visualise the database of already extracted information as a co-occurrence table together with its context.
Figure 9 illustrates the application technology stack used to implement the ADE Discovery Browser.
Examples of Retrieved Information
Figure 10 illustrates the first user scenario where the user can input a paragraph and visualise the predictions below having the identified entities highlighted and the relations listed on the table.
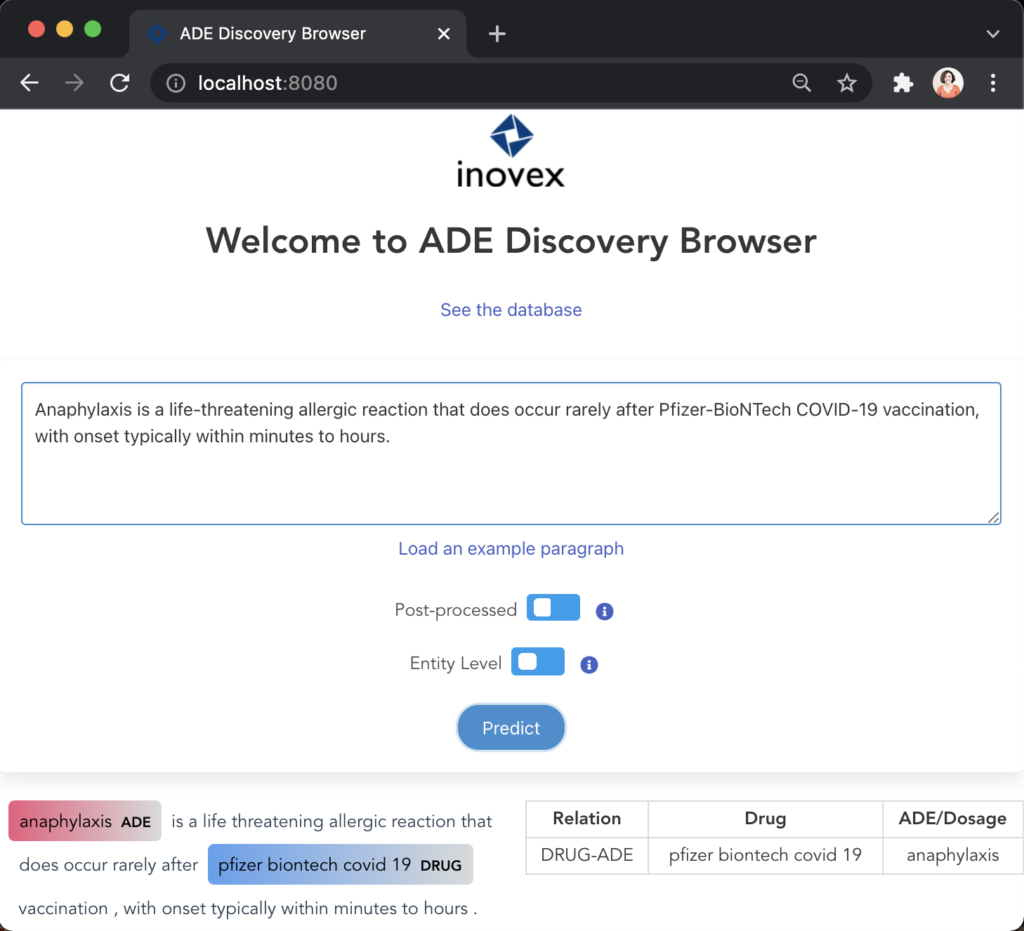
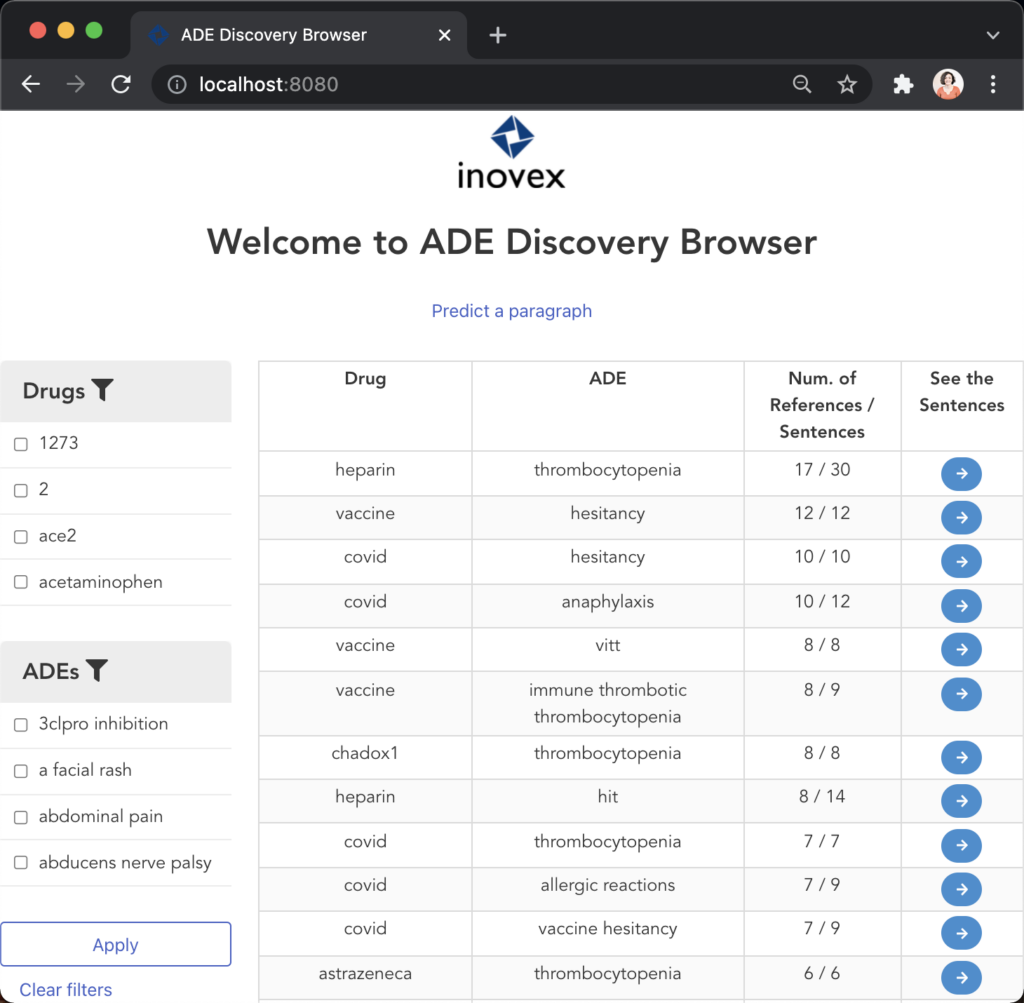
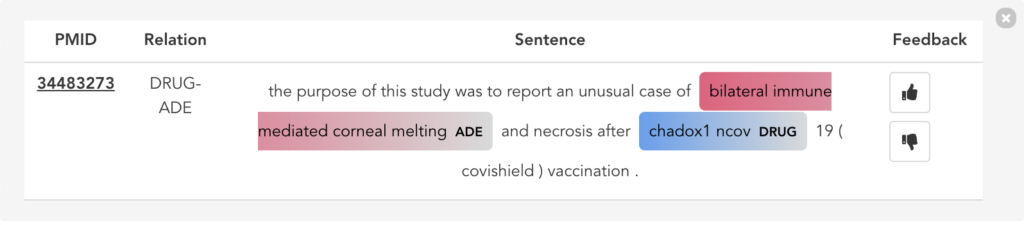
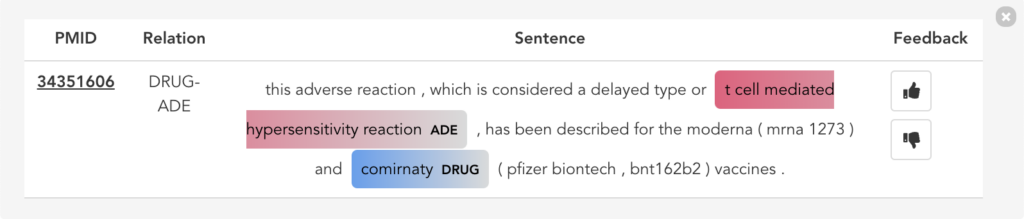
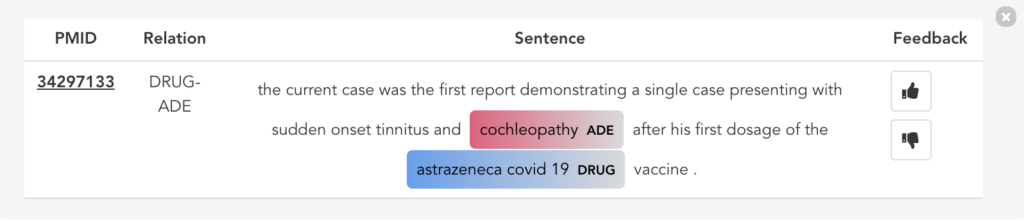
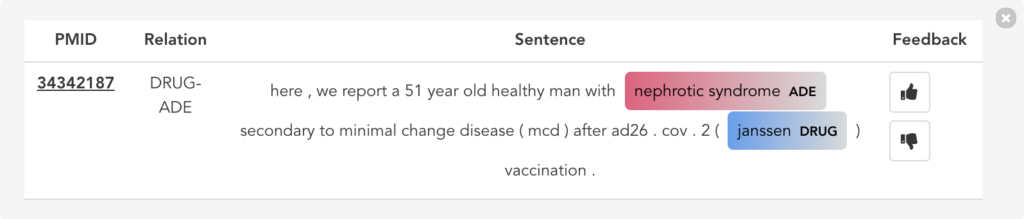
Figure 11 illustrates the second user scenario and lists the extracted DRUG-ADE tuples from PubMed aggregated as a co-occurrence table. Drug and ADE column refers to the recognised drug and ADE mentions respectively. Next to that, we can see the number of literature and the number of sentences the specific tuple was used. Because the table includes over a thousand tuples, it becomes difficult to find specific information. Therefore, we added drug and ADE filters on the left side. When the “See the Sentences“ button is clicked for a tuple, a pop-up window will be shown to visualise the context the tuple was used as illustrated in Figure 12.



Conclusion
ADE extraction using NLP techniques still did not reach its perfectly performing level due to the language variety in different domains and a lack of quality of the annotated data. Additionally, when applying machine learning in life sciences, human intervention is important to have the predictions reviewed by experts for correctness. Therefore, combining such AI based ADE extraction with a system where user interaction is possible is a good use case for this task. The idea behind the ADE Discovery Browser is therefore, to provide a user-friendly platform to the pharmacovigilance experts to scroll through the potential DRUG, ADE entities and the potential relations between them which are extracted from the PubMed. Eventually, the experts can approve or disapprove the extracted information seeing the sentence in which context the two target entities are potentially related to each other.
Considering the extracted information about the ADEs related with COVID-19 vaccines, we conclude that PLMs can be fine-tuned for ADE extraction incorporating outside and contextual knowledge to perform token and sentence classification for NER and RE respectively, and adequately be used for information retrieval from other similar domain text resources having just as good performing models as the best team, and few labeled data.
NOTE: This study was within the scope of Mürüvvet Hasanbaşoğlu’s master’s thesis study at TUM under the close supervision of Dr. Robert Pesch and Sebastian Blank, and has not been used for any other purpose.
References
[1] https://www.nlpsummit.org/adverse-drug-event-detection-using-spark-nlp/
[2] Edwards, I. Ralph, and Jeffrey K. Aronson. „Adverse drug reactions: definitions, diagnosis, and management.“ The lancet 356.9237 (2000): 1255-1259.
[3] Leape, Lucian L., et al. „The nature of adverse events in hospitalized patients: results of the Harvard Medical Practice Study II.“ New England journal of medicine 324.6 (1991): 377-384.
[4] Bates, David W., et al. „Incidence of adverse drug events and potential adverse drug events: implications for prevention.“ Jama 274.1 (1995): 29-34.
[5] Henry, Sam, et al. „2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records.“ Journal of the American Medical Informatics Association 27.1 (2020): 3-12.
[6] Gurulingappa, Harsha, et al. „Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports.“ Journal of biomedical informatics 45.5 (2012): 885-892.
[7] Wei, Qiang, et al. „A study of deep learning approaches for medication and adverse drug event extraction from clinical text.“ Journal of the American Medical Informatics Association 27.1 (2020): 13-21.
[8] Zhang, Yuhao, et al. „Position-aware attention and supervised data improve slot filling.“ Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing. 2017.



